{
"info": {
"author": "Brutenis Gliwa",
"author_email": "",
"bugtrack_url": null,
"classifiers": [
"License :: OSI Approved :: GNU General Public License v3 (GPLv3)",
"Programming Language :: Python",
"Programming Language :: Python :: 3",
"Programming Language :: Python :: 3.6",
"Programming Language :: Python :: Implementation :: CPython",
"Topic :: Scientific/Engineering :: Medical Science Apps."
],
"description": "\nAirway
\n\n\n \n  \n \n\n \n
\n \n\n \n  \n \n\n \n
\n \n\n \n  \n \n\n \n
\n \n\n \n  \n \n
\n \n
\n\nAnatomical classification of human lung bronchus up to segmental/tertiary bronchi \nbased on high-resolution computed tomography (HRCT) image masks.\nA rule-based approach is taken for classification, where the most cost-effective tree is found according\nto their angle deviations by defined vectors.\n\nHere, a pipeline is implemented which, given a formatted input data structure, can create the anatomical segmentations, clusters of similar anatomy, and\nthe visualisations presented below. \n\nAn example can be seen below, the 18 segments of the lung are automatically annotated in \nthe 3D voxel model.\n\n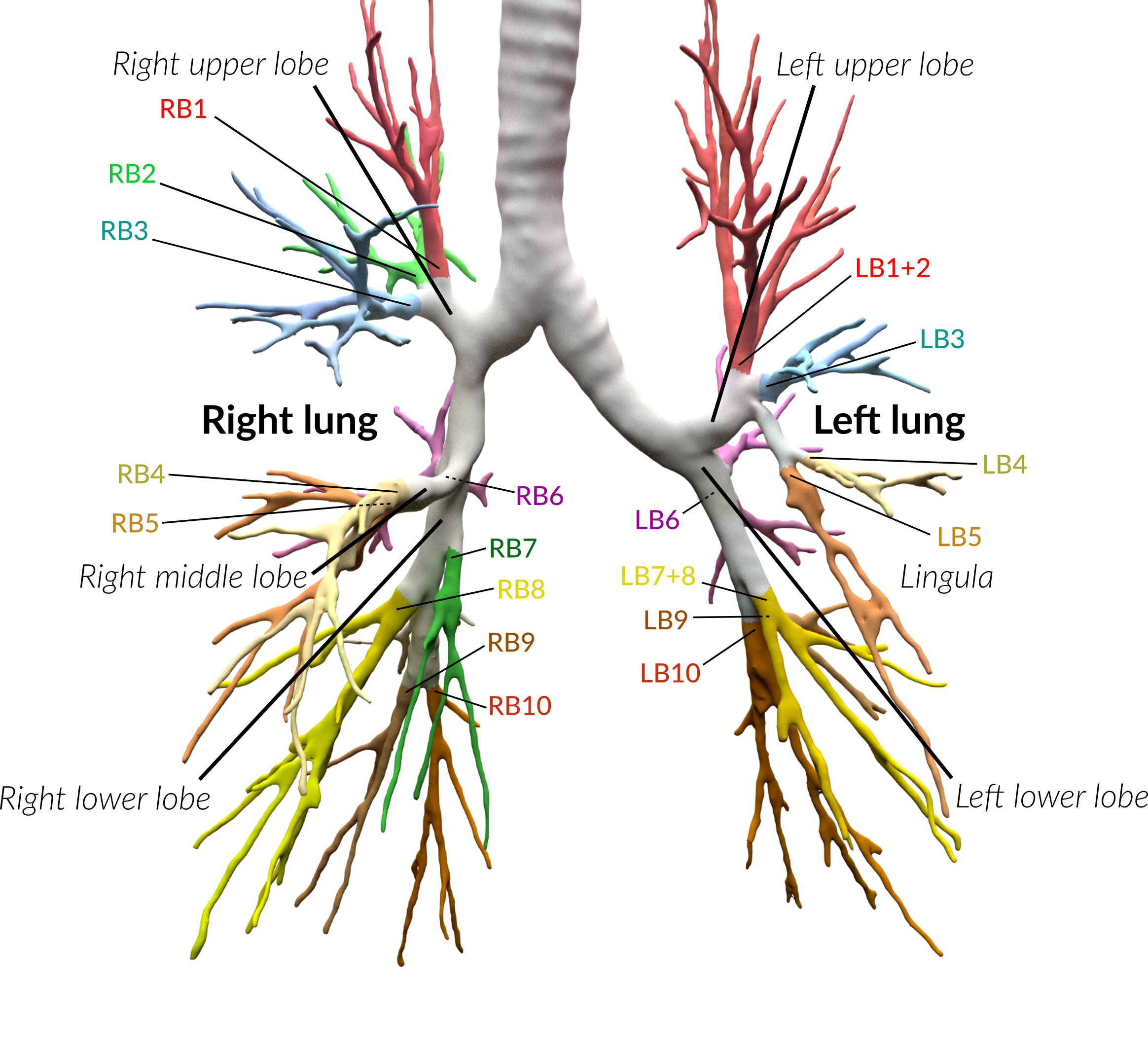\n\n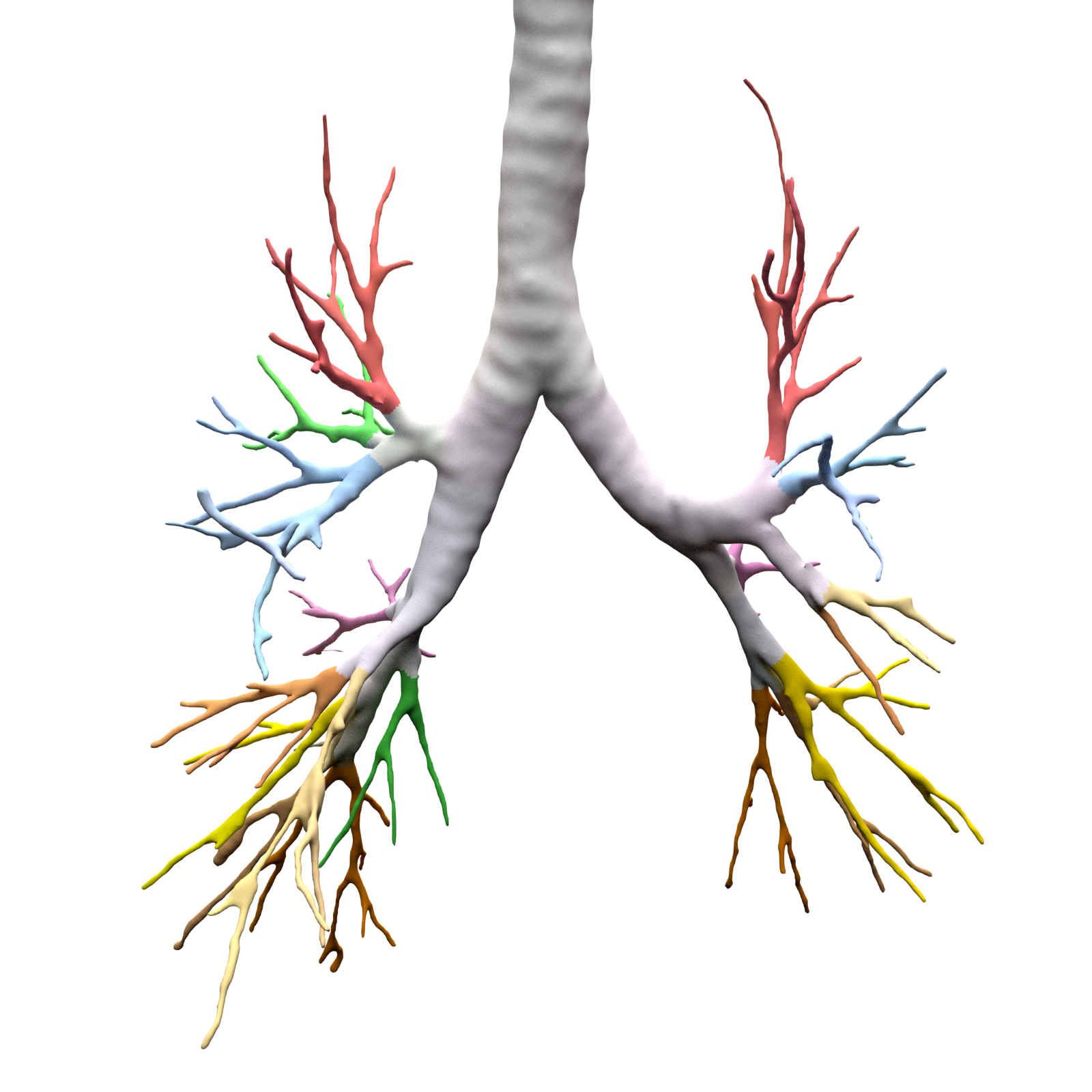\n\nExample visualisation of split detection, rendered with Blender.\n\nThis project currently uses masks created using Synapse 3D by Fujifilm, \nwhich already segments the lobes, bronchus, arteries and veins in CT images. \nHowever, this is not required. To use this project you only need a detailed segmentation of the bronchi.\n\n# Quickstart\n\n* Install with `pip install airway`\n* Run the interactive tutorial with `airway tutorial` (this will guide you through a fake sample)\n* Modify your own data so that you could use it with `airway` (described in the data section below)\n\n\n# Data\n\nWe use a pipeline based approach to calculate the raw data. With the help of\n`airway_cli.py` you are able to calculate each step (called stages).\n\nTo get the pipeline to work you need to define and format the first input stage, which is then\nused to create all other stages.\nYou have multiple options which stage you use as input, depending on which is simpler\nfor your use case: \n\n* `raw_data` is the structure as created by Synapse 3D and cannot be\ndirectly used as input. We used the script in `scripts/separate-bronchus-files.sh`\nto create `raw_airway`. The format of it is still described in the `DATADIR` graphic below for reference.\n* `raw_airway` is the same data as `raw_data`, but the directory structure has been reformatted. This was our\ninput stage for the pipeline. See the `DATADIR` graphic below for more details on the file structure.\n The `IMG\\d` files contain single slices for the CT scan, where -10000 was used for empty, and the rest \n were various values between -1000 to 1000. We assumed -10000 to be empty, and everything else to be\n a voxel of that type, as we already had segmented data.\n* `stage-01` **(recommended)** can be used as an input stage as well, this may be considerably easier to compute\nif you have a wildly different data structure. Only a single file needs to be created: `model.npz`. \n It is a compressed numpy file where a single ~800\u00d7512\u00d7512 array is saved for the entire lung \n (order is important, (_transverse_ \u00d7 _sagittal_ \u00d7 _frontal_ planes)). \nThe ~800 is variable and depends on the patient, the 512\u00d7512 is the slice dimension.\n The array is of type `np.int8` and\n the array in the `.npz` file is not named (it should be accessible as `arr_0`). \n An encoding is used for each voxel to represent the 8 different classes as shown in the table below. \n If you do not have some classes then you may ignore them, only `bronchus` (encoded as `1`) is required,\n as otherwise nothing will really work in the rest of the project. Empty or air should be encoded as `0`.\n See `airway/image_processing/save_images_as_npz.py` for reference if you decide to use this stage as input.\n\n \nNote that the slice thickness for our data was 0.5 mm in all directions. \nCurrently, the pipeline assumes this is always the case. \nIt will work fairly well for different, but equal, thicknesses in all directions (e.g. 0.25 mm \u00d7 0.25 mm \u00d7 0.25 mm), \nalthough some results may wary. \nDifferent thicknesses in multiple directions (e.g. 0.8 mm \u00d7 0.8 mm \u00d7 3 mm) will likely not work well at all. In that case\nwe recommend to duplicate certain axes manually, so that the thickness is similar in all directions.\n\n\n| Category | Encoding |\n| --- | --- |\n| Empty | 0 |\n| Bronchus | 1 |\n| LeftLowerLobe | 2 |\n| LeftUpperLobe | 3 |\n| RightLowerLobe | 4 |\n| RightMiddleLobe | 5 |\n| RightUpperLobe | 6 |\n| Vein | 7 |\n| Artery | 8 |\n\nThe directory structure for the data structure is described below. Note that if you use `stage-01` as input you do not need \n`raw_data` or `raw_airway` at all.\n\n```\n DATADIR\n \u251c\u2500\u2500 raw_data \ud83e\udc14 This is an example of entirely unformatted raw data as we received them\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 Ct_Thorax_3123156 \ud83e\udc14 Each patient has its own directory \n \u2502\u00a0\u00a0 \u2514\u2500\u2500 DATA\n \u2502\u00a0\u00a0 \u00a0\u00a0 \u251c\u2500\u2500 Data.txt \ud83e\udc14 This contained the paths for finding the various bronchus and lobes\n \u2502\u00a0\u00a0 \u00a0\u00a0 \u2514\u2500\u2500 3123156 \ud83e\udc14 Example patient ID\n \u2502\u00a0\u00a0 \u00a0\u00a0 \u2514\u2500\u2500 20190308 \n \u2502\u00a0\u00a0 \u00a0\u00a0 \u2514\u2500\u2500 124101 \n \u2502\u00a0\u00a0 \u00a0\u00a0 \u2514\u2500\u2500 EX1 \n \u2502\u00a0\u00a0 \u00a0\u00a0 \u251c\u2500\u2500 SE1 \ud83e\udc14 Each SE* folder contains a list of DICOM images \n \u2502\u00a0\u00a0 \u00a0\u00a0 \u2502 \u251c\u2500\u2500 IMG1 named IMG1 through IMG642 (may be a different amount)\n \u2502\u00a0\u00a0 \u00a0\u00a0 \u2502 \u251c\u2500\u2500 ... these represent the slices for that segmentation.\n \u2502\u00a0\u00a0 \u00a0\u00a0 \u2502 \u2514\u2500\u2500 IMG642 E.g. SE4 is Bronchus, SE5 is the Right upper lobe.\n \u2502\u00a0\u00a0 \u00a0\u00a0 \u251c\u2500\u2500 SE2 This is described in Data.txt for each patient.\n \u2502\u00a0\u00a0 \u00a0\u00a0 \u251c\u2500\u2500 ... \n \u2502\u00a0\u00a0 \u00a0\u00a0 \u2514\u2500\u2500 SE10 \n \u2502\u00a0\u00a0 \u00a0\u00a0 \u2514\u2500\u2500 SE11 \n \u2502\u00a0\u00a0 \u00a0\u00a0 \n \u251c\u2500\u2500 raw_airway \ud83e\udc14 Formatted data which will be used as input for stage-01\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 3123156 \ud83e\udc14 Single patient folder, in total there are around 100 of these\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 Artery\n \u2502\u00a0\u00a0 \u2502 \u251c\u2500\u2500 IMG1 \ud83e\udc14 DICOM images, in our case 512x512 slices\n \u2502\u00a0\u00a0 \u2502 \u251c\u2500\u2500 IMG2 \ud83e\udc14 with 0.5 mm thickness in all directions\n \u2502\u00a0\u00a0 \u2502 \u251c\u2500\u2500 ...\n \u2502\u00a0\u00a0 \u2502 \u251c\u2500\u2500 IMG641 \ud83e\udc14 There generally are between 400 and 800 of these slices\n \u2502\u00a0\u00a0 \u2502 \u2514\u2500\u2500 IMG642 \ud83e\udc14 So the number of slices is variable\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 Bronchus\n \u2502\u00a0\u00a0 \u2502 \u251c\u2500\u2500 IMG1 \ud83e\udc14 Same number and dimension of slices as above\n \u2502\u00a0\u00a0 \u2502 \u251c\u2500\u2500 ...\n \u2502\u00a0\u00a0 \u2502 \u2514\u2500\u2500 IMG642 \n \u2502\u00a0\u00a0 \u251c\u2500\u2500 LeftLowerLobe \ud83e\udc14 All of these also share the same structure\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 LeftUpperLobe\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 RightLowerLobe\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 RightMiddleLobe\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 RightUpperLobe\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 Vein\n \u2502\u00a0\u00a0 \u00a0\u00a0 \n \u251c\u2500\u2500 stage-01 \ud83e\udc14 Each stage now has the same basic format\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 3123156\n \u2502\u00a0\u00a0 \u2502 \u2514\u2500\u2500 model.npz \ud83e\udc14 See above for an explanation\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 3123193\n \u2502\u00a0\u00a0 \u2502 \u2514\u2500\u2500 model.npz\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 ...\n \u251c\u2500\u2500 stage-02 \ud83e\udc14 Each stage from here on will be created by the pipeline itself\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 3123156 so you do not need to handle this, each of them have different\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 ... files depending on their use.\n ...\n\n```\n\nNote that currently NIFTI images are not supported, all `IMG\\d` files are DICOM images.\n\n\n\n# Installation\n\nAt least Python 3.6 is required for this project.\n\n```\npip3 install airway\n```\n\nThe open source 3D visualisation software [Blender](https://www.blender.org/) is required for visualisation. This dependency is optional if you do not need the visualisation part. Install from the website above or via a package manager like this (pip does not have blender): \n\n```apt install blender```\n\nTested with Blender versions 2.76, 2.79, 2.82 (recommended) and 2.92. \n\nNow configure the defaults, copy and rename `configs/example_defaults.yaml` to `configs/defaults.yaml` \n(in the root folder of the project) and change the path in the file to where you have put the data.\nYou may ignore the other parameters for now, although feel free to read the comments there and adjust\nthem as needed (especially number of workers/threads).\n\n\n\n# Stages\n\nFor every calculated stage `airway` creates a new directory (`stage-xx`) and \nsubdirectories for each patient (e.g. `3123156`).\n\nEach stage has input stages, these are handled for you though, so you only need to specify which stages to create.\nIf you use `raw_airway` as input stage, then calculate `stage-01`:\n\n airway stages 1\n\nYou may add the `-1` flag to calculate a single patient for test purposes. Note that calculation of `stage-01` \nmay be really slow if you store your data on an HDD (multiple hours), as there are a lot of single small files with a large\noverhead for switching between files. \n\nOr if you use `stage-01` as input you can calculate `stage-02` directly:\n\n airway stages 2\n\nIf this works then great! You may continue to create all other stages as described below. \nIf it does not work, then make sure the data format is correct as described in the **Data** section. \nIf you think everything is correct then please open an issue or message me, there may be bugs, or some stuff\nmay be naively hard-coded.\n\nYou may list all stages with short descriptions by calling `airway stages` without any arguments,\nor you can list all commands by using the `--help` flag.\n\nSummary of the stages:\n\n* Stages **1 - 7** use the raw data to create the tree splits used in the rest of the stages.\n* Stages **30 - 35** analyse the tree structure, focusing mostly on the left upper lobe.\n* Stages **60 - 62** are 3D visualisations, wherein .obj files of the lungs are exported.\n* Stages **70 - 72** are plot visualisations of various stats.\n* Stage **90** is the website which displays information for each patient including the 3D models.\n\nThe airway pipeline checks if the stage already exists, if you need to overwrite\na stage you need to add the `-f`/`--force` flag.\n\nYou can now create all remaining stages like this:\n\n airway stages 2+\n\n\nIt may take a couple of hours for everything, depending on how many patients you have.\nIf you don't have some dependencies installed you can still safely run it, and only those stages will crash.\nOpen the `./log` file and search for `STDERR` if you want to see the errors listed by `airway`.\n\nBy default, eight patients will be calculated in parallel (8 workers are used).\nIf you have more CPU threads, simply increase the number of workers:\n\n`airway stages 1 2 3 -w 32` or change the default in the config file (`defaults.yaml`).\n\nTo see the results you may open blender interactively like this:\n\n`airway vis 1 -o`\n\nThis loads the bronchus model with the correct materials for the segments.\n\nYou can also see the various files created by the stages:\n* `stage-62`: renders based on the lung\n* `stage-10`: which contain `.graphml` files describing the tree structure, \n and the classifications created by the algorithm.\n* `stage-35`: creates a pdf with renders for each patient\n* `stage-11`: creates a pdf with the found clusters of the various structures \n\n# More images\n\n\n\n\n\n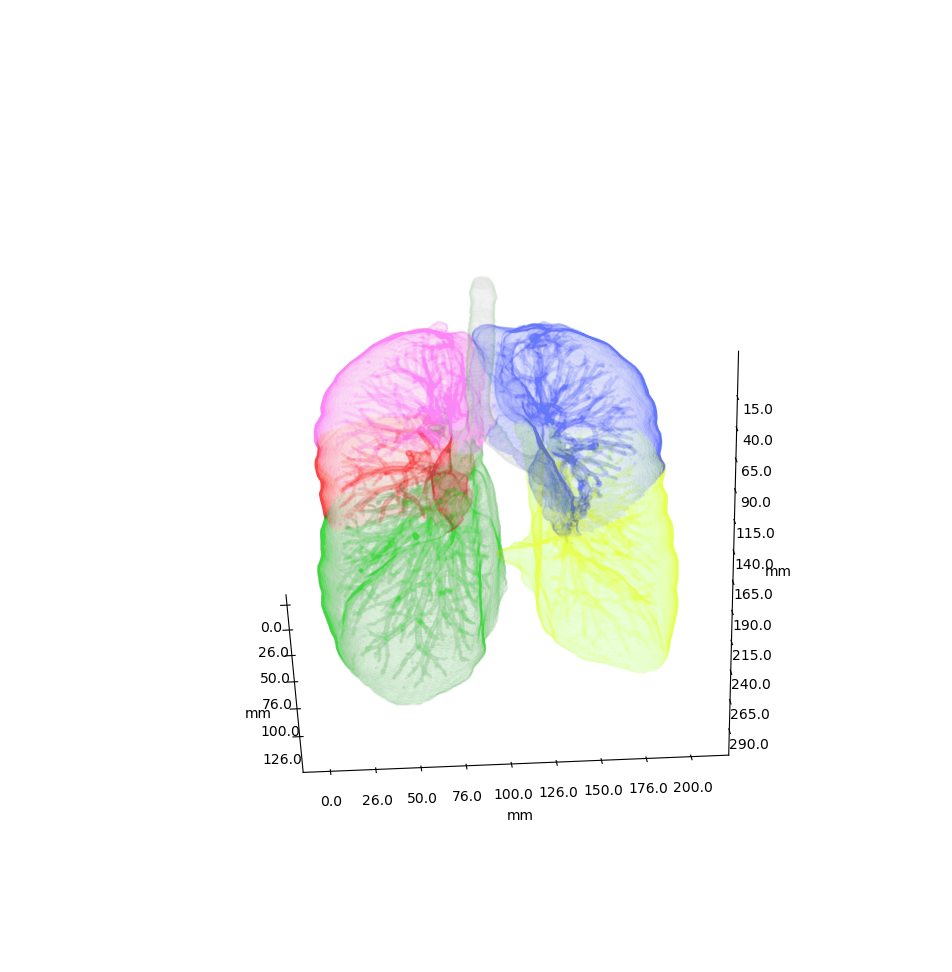\n\n## Example trees for patient 3183090\n\n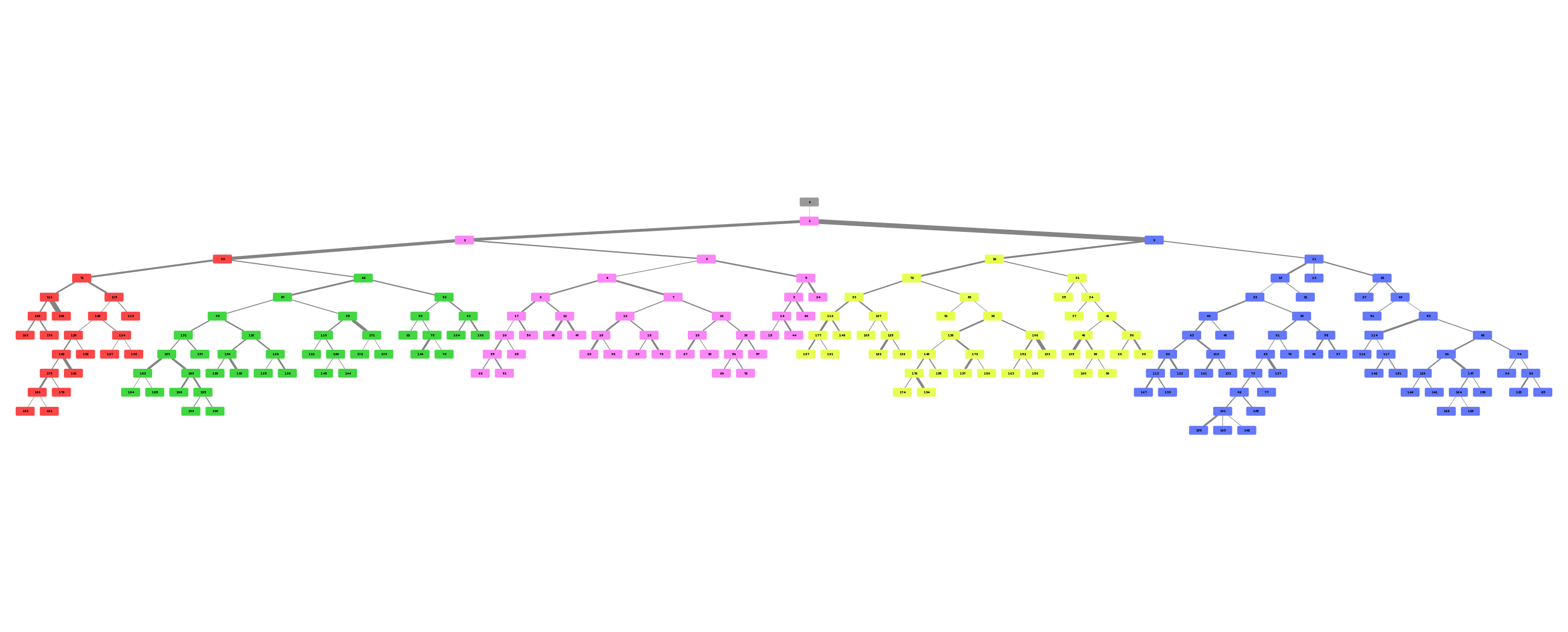\n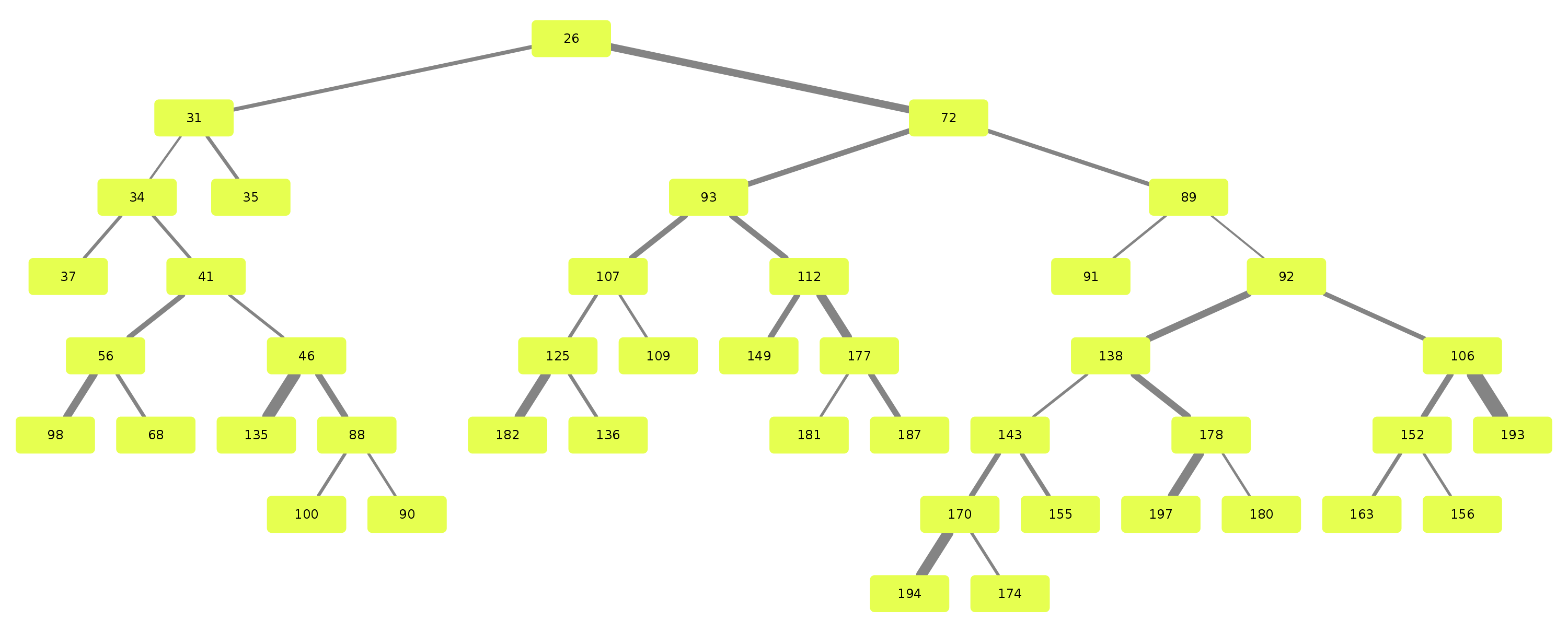\n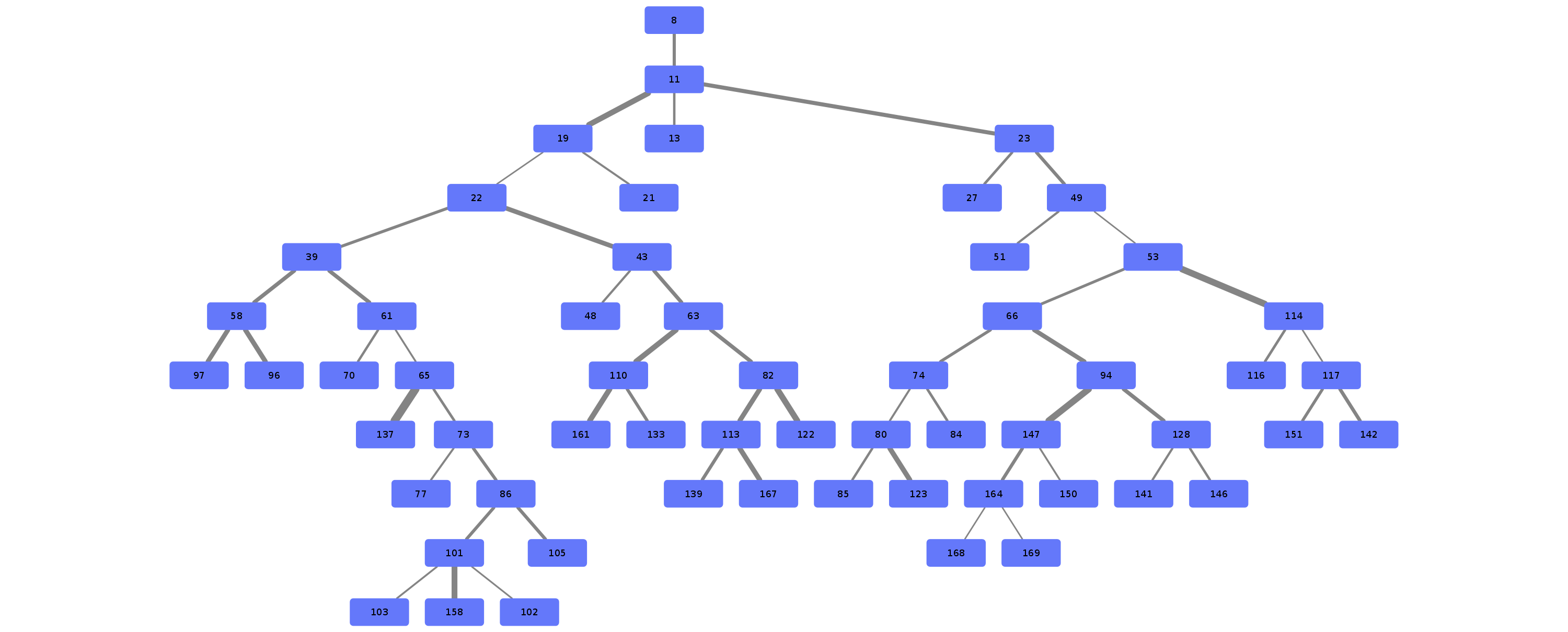\n\n\n\n\n# Credits & Thanks to\n\nAirway originated as an observation by Dr. Rolf Oerter at the University of Rostock\nthat certain structures in the lungs bronchus he has seen while operating have not been documented. \nThe first steps of the project were made as a student project at the University of Rostock\nat the Department of Systems Biology organised by Mariam Nassar. \n\nIt consisted of this team:\n\n- [Martin Steinbach](https://github.com/meetunix)\n- [Brutenis Gliwa](https://github.com/liquidfun)\n- Lukas Gro\u00dfehagenbrock\n- Jonas Moesicke\n- Joris Thiele\n\nAfter this, the project is being continued by me (Brutenis) as my bachelor thesis. \nThanks to Mariam Nassar, Dr. Rolf Oerter, Gundram Leifert and Prof. Olaf Wolkenhauer for supervision during this time.\nAnd thanks to Planet AI for letting me write my thesis at their office.\n\n",
"description_content_type": "text/markdown",
"docs_url": null,
"download_url": "",
"downloads": {
"last_day": -1,
"last_month": -1,
"last_week": -1
},
"home_page": "https://github.com/LiquidFun/Airway",
"keywords": "",
"license": "GPL-3.0",
"maintainer": "",
"maintainer_email": "",
"name": "airway",
"package_url": "https://pypi.org/project/airway/",
"platform": null,
"project_url": "https://pypi.org/project/airway/",
"project_urls": {
"Homepage": "https://github.com/LiquidFun/Airway"
},
"release_url": "https://pypi.org/project/airway/0.2.3/",
"requires_dist": [
"numpy",
"pydicom",
"networkx",
"scikit-image",
"pandas",
"pyyaml",
"matplotlib",
"tqdm",
"Markdown",
"weasyprint",
"pytest",
"py2cytoscape"
],
"requires_python": ">=3.6",
"summary": "Automatic classification of tertiary bronchi based on bronchus masks using a rule-based approach.",
"version": "0.2.3",
"yanked": false,
"yanked_reason": null
},
"last_serial": 15705799,
"releases": {
"0.1.0": [
{
"comment_text": "",
"digests": {
"md5": "1bf84628a31cdbeba3a50f02d0a4ca12",
"sha256": "0ccc2efd860310b4bd278afb3c180e05e271e2297b19be0438bcfb87c6cfde57"
},
"downloads": -1,
"filename": "airway-0.1.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "1bf84628a31cdbeba3a50f02d0a4ca12",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.6.0",
"size": 18672,
"upload_time": "2021-05-31T17:41:55",
"upload_time_iso_8601": "2021-05-31T17:41:55.999514Z",
"url": "https://files.pythonhosted.org/packages/68/ed/f66c5bd837671a528aa3b6f8fafaf928a8d39c0f921616c5ea604df53081/airway-0.1.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"md5": "7f450c8e5206b0e0ecfdfb121dda6645",
"sha256": "c70572db7a2dfb20902ab0f89f1760219149196e7e83ae088bb45a5e57ba5d56"
},
"downloads": -1,
"filename": "airway-0.1.0.tar.gz",
"has_sig": false,
"md5_digest": "7f450c8e5206b0e0ecfdfb121dda6645",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6.0",
"size": 20755,
"upload_time": "2021-05-31T17:41:57",
"upload_time_iso_8601": "2021-05-31T17:41:57.497435Z",
"url": "https://files.pythonhosted.org/packages/38/28/f8d6e6f2cde428d3a82588213d996820a84562172642feffd7fdce45514f/airway-0.1.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"0.1.1": [
{
"comment_text": "",
"digests": {
"md5": "5eb703667542cfb1ac799744e9a00748",
"sha256": "fb60d6504e477173b4950950600858cbf734eab31735bffe8b26267c6197a276"
},
"downloads": -1,
"filename": "airway-0.1.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "5eb703667542cfb1ac799744e9a00748",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.6",
"size": 18723,
"upload_time": "2021-05-31T17:59:12",
"upload_time_iso_8601": "2021-05-31T17:59:12.762082Z",
"url": "https://files.pythonhosted.org/packages/cf/34/97753045e362786856e73f7143cf4b458e3baaa875690fd8327734d5de69/airway-0.1.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"md5": "dbb7d973d851014cfa28302040413a7e",
"sha256": "2589403ac6e696b1b65bbc00dcaf27632e70a2117beee22fa891bb8e0542dfd9"
},
"downloads": -1,
"filename": "airway-0.1.1.tar.gz",
"has_sig": false,
"md5_digest": "dbb7d973d851014cfa28302040413a7e",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6",
"size": 20797,
"upload_time": "2021-05-31T17:59:13",
"upload_time_iso_8601": "2021-05-31T17:59:13.699499Z",
"url": "https://files.pythonhosted.org/packages/1e/d6/d9075422e69528a43604eab4e9c9deaf9fd5fa648110d57bcd4e9cb300d5/airway-0.1.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"0.2.0": [
{
"comment_text": "",
"digests": {
"md5": "468580f368b290425acf8739cb6d14da",
"sha256": "152f7315281bdb6223ba228e3de5cf4b5ab8a35edde4395dabc518548e5ca758"
},
"downloads": -1,
"filename": "airway-0.2.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "468580f368b290425acf8739cb6d14da",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.6",
"size": 99134,
"upload_time": "2022-10-18T09:58:45",
"upload_time_iso_8601": "2022-10-18T09:58:45.326439Z",
"url": "https://files.pythonhosted.org/packages/82/01/febe9bf8db7e3967dd0c1f6060147bbd22a74b9e673abf47950b818ceaff/airway-0.2.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"md5": "e96d02397a534642dc49a8a90272b881",
"sha256": "a0825b93ad5aab4a7a8f0b0e294841511153c78a1c7b99545c2adbb308503d5d"
},
"downloads": -1,
"filename": "airway-0.2.0.tar.gz",
"has_sig": false,
"md5_digest": "e96d02397a534642dc49a8a90272b881",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6",
"size": 84262,
"upload_time": "2022-10-18T09:58:46",
"upload_time_iso_8601": "2022-10-18T09:58:46.636640Z",
"url": "https://files.pythonhosted.org/packages/85/f3/a26f610b1cae858c32987215c4e01b6057025ce8a910c6375274ad895af3/airway-0.2.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"0.2.1": [
{
"comment_text": "",
"digests": {
"md5": "0aed34fca2a4f5ff6546f36859c0fcbf",
"sha256": "e2955f3456ee9e124941f59b0af212f7763cf19857dfbc5f686f70294714b2a4"
},
"downloads": -1,
"filename": "airway-0.2.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "0aed34fca2a4f5ff6546f36859c0fcbf",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.6",
"size": 254315,
"upload_time": "2022-11-06T01:39:46",
"upload_time_iso_8601": "2022-11-06T01:39:46.945189Z",
"url": "https://files.pythonhosted.org/packages/10/02/20d8c6c921611c1c1545b88d6be3b40712fa15bd4c8155bb0c8a758ff74a/airway-0.2.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"md5": "60788e32cb7021d1b157729ea100c1ee",
"sha256": "0d667340032a8f9275873c03555619c56221b87757fbf2683f93e74376a6b243"
},
"downloads": -1,
"filename": "airway-0.2.1.tar.gz",
"has_sig": false,
"md5_digest": "60788e32cb7021d1b157729ea100c1ee",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6",
"size": 240004,
"upload_time": "2022-11-06T01:39:48",
"upload_time_iso_8601": "2022-11-06T01:39:48.817932Z",
"url": "https://files.pythonhosted.org/packages/6e/18/6b693e47215835e2b0c932853441840cf87959a3af38a2580d7bd3d7ef29/airway-0.2.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"0.2.3": [
{
"comment_text": "",
"digests": {
"md5": "b67cd2ee76de417a9bf1adf73f740ddd",
"sha256": "1a272f8ccddfe7aeb9c8c3d18ec9046e353082cf0e27823272d5553e443fe989"
},
"downloads": -1,
"filename": "airway-0.2.3-py3-none-any.whl",
"has_sig": false,
"md5_digest": "b67cd2ee76de417a9bf1adf73f740ddd",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.6",
"size": 254490,
"upload_time": "2022-11-09T00:57:07",
"upload_time_iso_8601": "2022-11-09T00:57:07.888895Z",
"url": "https://files.pythonhosted.org/packages/17/8a/f00ff1dfb87be843997406f8e899a40b9d0c3cabe816620bdd96e83eb11c/airway-0.2.3-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"md5": "7f9f25c7c3bb154c4f13a1d3fe147e33",
"sha256": "b9ff4f0453cd584460017c779468054f421c8586d86ffdba8169cdf993a1af8c"
},
"downloads": -1,
"filename": "airway-0.2.3.tar.gz",
"has_sig": false,
"md5_digest": "7f9f25c7c3bb154c4f13a1d3fe147e33",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6",
"size": 240135,
"upload_time": "2022-11-09T00:57:09",
"upload_time_iso_8601": "2022-11-09T00:57:09.544435Z",
"url": "https://files.pythonhosted.org/packages/e3/2d/244b8408f6a45406ab79d0de9dfdb9e0358a08a37c737a829357c6b1c703/airway-0.2.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
]
},
"urls": [
{
"comment_text": "",
"digests": {
"md5": "b67cd2ee76de417a9bf1adf73f740ddd",
"sha256": "1a272f8ccddfe7aeb9c8c3d18ec9046e353082cf0e27823272d5553e443fe989"
},
"downloads": -1,
"filename": "airway-0.2.3-py3-none-any.whl",
"has_sig": false,
"md5_digest": "b67cd2ee76de417a9bf1adf73f740ddd",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.6",
"size": 254490,
"upload_time": "2022-11-09T00:57:07",
"upload_time_iso_8601": "2022-11-09T00:57:07.888895Z",
"url": "https://files.pythonhosted.org/packages/17/8a/f00ff1dfb87be843997406f8e899a40b9d0c3cabe816620bdd96e83eb11c/airway-0.2.3-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"md5": "7f9f25c7c3bb154c4f13a1d3fe147e33",
"sha256": "b9ff4f0453cd584460017c779468054f421c8586d86ffdba8169cdf993a1af8c"
},
"downloads": -1,
"filename": "airway-0.2.3.tar.gz",
"has_sig": false,
"md5_digest": "7f9f25c7c3bb154c4f13a1d3fe147e33",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6",
"size": 240135,
"upload_time": "2022-11-09T00:57:09",
"upload_time_iso_8601": "2022-11-09T00:57:09.544435Z",
"url": "https://files.pythonhosted.org/packages/e3/2d/244b8408f6a45406ab79d0de9dfdb9e0358a08a37c737a829357c6b1c703/airway-0.2.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"vulnerabilities": []
}  \n \n\n \n
\n \n\n \n  \n \n
\n \n